Extended-spectrum β-lactamases (ESBLs) represent a significant challenge in the fight against antibiotic resistance. Among these, the CTX-M family has become globally prevalent. This study focuses on the emergence of CTX-M-producing Enterobacteriaceae in French hospitals, with a particular emphasis on the CTX-M-3 variant. CTX-M-3 is a notable enzyme within this family due to its efficient hydrolysis of cefotaxime and its role in mediating resistance to crucial cephalosporin antibiotics.
The CTX-M enzymes are a class A type of ESBL, gaining prominence in the 1990s with the initial characterization of CTX-M-1 (MEN-1). These enzymes are distinguished by their enhanced hydrolytic activity against cefotaxime compared to ceftazidime, leading to high resistance to cefotaxime but having less impact on ceftazidime minimum inhibitory concentrations (MICs). The CTX-M family is diverse, encompassing at least twelve members categorized into four primary phylogenetic groups based on amino acid sequence similarities: the CTX-M-1 branch (including CTX-M-1, CTX-M-3, and CTX-M-10), the CTX-M-2 branch, the CTX-M-9 branch, and the CTX-M-8 branch. These enzymes have been identified in various Enterobacteriaceae species and even Vibrio cholerae, with significant geographical distribution across South America, Eastern Europe, and Japan.
This investigation details six clinical Enterobacteriaceae strains (detailed in Table 1) collected from different French hospitals. These strains were selected based on positive synergy test outcomes and higher resistance to cefotaxime than ceftazidime, with MICs ranging from 16 to 128 μg/ml and 2 to 8 μg/ml, respectively. Among these isolates, Enterobacter cloacae Ver-1 and CF-2 were identified as producing CTX-M-3 and CTX-M-1 respectively, while the remaining four strains produced CTX-M-14. Reference strains, including CTX-M-1 producer MEN and CTX-M-3-encoding plasmid A1, were utilized for comparative analysis.
Analytical isoelectric focusing revealed that all tested strains produced TEM-1 penicillinase (pI 5.4), along with a second β-lactamase of alkaline pI. Specifically, strains Ver-1 and CF-2 exhibited a pI of 8.4, consistent with CTX-M-1, while strains CF-1, Mnt-1, Mnt-2, and Roa-1 showed a pI of 7.9, corresponding to CTX-M-14. PCR and sequencing confirmed the presence of TEM-1. Further amplification using CTX-M specific primers yielded positive results, indicating the presence of blaCTX-M genes. Sequencing of the blaCTX-M genes from strains Ver-1 and CF-2 confirmed 100% identity to blaCTX-M-3 and blaCTX-M-1, respectively. The blaCTX-M gene from strain CF-1 was cloned and sequenced, revealing it to be blaCTX-M-14, consistent with findings for strains Mnt-1, Mnt-2, and Roa-1 which also harbored blaCTX-M-14.
The deduced amino acid sequence of CTX-M-14 differed from CTX-M-9 by a single amino acid substitution, Ala-231→Val. Kinetic studies of purified CTX-M-14 enzyme showed similar kinetic constants to CTX-M-9, with both enzymes demonstrating high catalytic efficiency against various penicillins and cephalosporins, but lower activity against ceftazidime and aztreonam. Both CTX-M-14 and CTX-M-9 were susceptible to β-lactamase inhibitors like clavulanate and tazobactam.
Conjugation experiments confirmed the transferability of cefotaxime resistance mediated by these CTX-M enzymes. MIC testing of transconjugants demonstrated high resistance to amoxicillin, ticarcillin, cephalothin, and cefuroxime. Cefotaxime MICs were significantly higher than ceftazidime MICs, and clavulanate effectively restored the activity of β-lactams.
Plasmid analysis (Figure 1A) revealed that CTX-M enzymes were encoded on plasmids of varying sizes: 55-kb plasmid pCF-2 (CTX-M-1), 180-kb plasmid pVer-1 (CTX-M-3), 150-kb plasmid pCF-1 (CTX-M-14), 110-kb plasmid pMnt-1 (CTX-M-14), and 120-kb plasmid pRoa-1 (CTX-M-14). Restriction enzyme digestion (Figure 1B) showed distinct patterns for plasmids encoding different CTX-M types, although pRoa-1 and pMnt-1 (both CTX-M-14 encoding) exhibited related patterns. Notably, CTX-M-1-encoding plasmid pCF-2 and CTX-M-3-encoding plasmid pVer-1 showed relatedness to previously reported plasmids pMEN and A1, respectively, suggesting plasmid diffusion across different geographical locations and time periods.
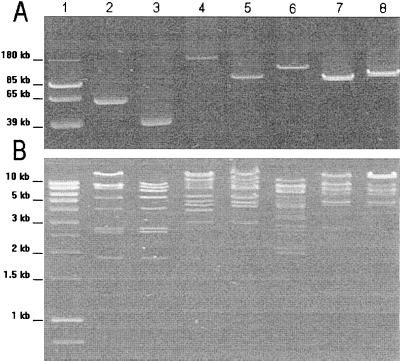 Agarose gel electrophoresis of plasmid DNA from CTX-M-producing E. coli transconjugants, illustrating plasmid sizes and bands.
Agarose gel electrophoresis of plasmid DNA from CTX-M-producing E. coli transconjugants, illustrating plasmid sizes and bands.
Figure 1: (A) Agarose gel electrophoresis showing plasmid DNA from CTX-M-producing E. coli transconjugants. (B) Agarose gel electrophoresis of HpaI-digested plasmid DNA. Lane details are provided in the original figure legend.
Further investigation revealed the presence of the ISEcp-1 element upstream of the blaCTX-M genes in all plasmids examined. This finding supports the hypothesis that ISEcp-1 plays a role in the mobilization and expression of blaCTX-M genes. The close proximity of ISEcp-1 to the blaCTX-M genes, located only 43 bp upstream, may contribute to enhanced blaCTX-M gene expression.
This study highlights the early emergence and dissemination of CTX-M-1, CTX-M-3, and CTX-M-14 ESBLs within French hospitals. The genetic context, particularly the association with ISEcp-1 and plasmid-mediated carriage, underscores the mechanisms driving the spread of these critical antibiotic resistance determinants. The presence of CTX-M-3, alongside other variants, emphasizes the ongoing evolution and dissemination of CTX-M-type ESBLs, posing a continued threat to public health and necessitating ongoing surveillance and control efforts.
References (Same as original article – not included here for brevity but would be listed in a full article).
TABLE 1. Clinical strains and plasmids used in the study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Clinical strains | ||
| E. coli CF-1 | Clinical strain harboring natural plasmid pCF-1 (Clermont-Ferrand, France, 1999) | This study |
| E. cloacae CF-2 | Clinical strain harboring natural plasmid pCF-2 (Clermont-Ferrand, France, 2000) | This study |
| E. coli Roa-1 | Clinical strain harboring natural plasmid pRoa-1 (Roanne, France, 1999) | This study |
| E. coli Mnt-1 | Clinical strain harboring natural plasmid pMnt-1 (Montpellier, France, 1999) | This study |
| K. pneumoniae Mnt-2 | Clinical strain producing CTX-M-14 (Montpellier, France, 1999) | This study |
| E. cloacae Ver-1 | Clinical strain harboring natural plasmid pVer-1 (Versailles, France, 1999) | 10 |
| E. coli MEN | Clinical strain harboring natural plasmid pMEN (Paris, France, 1999) | 2 |
| Plasmids | ||
| pCF-1 | 150-kb natural plasmid from E. coli strain CF-1 containing the blaCTX-M-14 gene | This study |
| pCF-2 | 55-kb natural plasmid from E. cloacae strain CF-2 containing the blaCTX-M-1 gene | This study |
| pMnt-1 | 110-kb natural plasmid from E. coli strain Mnt-1 containing the blaCTX-M-14 gene | This study |
| pRoa-1 | 120-kb natural plasmid from E. coli strain Roa-1 containing the blaCTX-M-14 gene | This study |
| pVer-1 | 180-kb natural plasmid from E. cloacae strain Ver-1 containing the blaCTX-M-3 gene | 10 |
| pMEN | 40-kb natural plasmid from E. coli strain MEN containing the blaCTX-M-1 gene | 2 |
| A1 | 110-kb natural A1 plasmid containing the blaCTX-M-3 gene | 20 |
| pClCF-1 | 13-kb recombinant plasmid of pBK-CMV containing the blaCTX-M-14 gene | This study |
| pBK-CMV | Phagemid vector; kanamycin resistance phenotype | Stratagene, La Jolla, Calif. |
TABLE 2. Substrate profile of CTX-M-14 and CTX-M-9 β-lactamases
| Substrate | CTX-M-14 | CTX-M-9 |
|---|---|---|
| *k*cat (s−1) | *K*m (μM) | |
| Penicillin G | 290 | 20 |
| Amoxicillin | 100 | 20 |
| Ticarcillin | 45 | 24 |
| Piperacillin | 200 | 48 |
| Cephalothin | 2,700 | 175 |
| Cefuroxime | 320 | 40 |
| Cefotaxime | 415 | 130 |
| Cefpirome | 940 | 1,000 |
| Aztreonam | 10 | 200 |
| Ceftazidime | 3 | 610a |
TABLE 3. Comparison of β-lactam MICs for CTX-M-producing E. coli transconjugants
| MIC (μg/ml) for E. coli C600 with plasmid (pI): |
|---|
| pCF-2a (5.4, 8.4) |
| Amoxicillin |
| Amoxicillin + CLAf |
| Ticarcillin |
| Ticarcillin + CLA |
| Piperacillin |
| Piperacillin + TZBg |
| Cephalothin |
| Cephalothin + CLA |
| Cefuroxime |
| Cefuroxime + CLA |
| Cefotaxime |
| Cefotaxime + CLA |
| Cefpirome |
| Cefpirome + CLA |
| Ceftazidime |
| Ceftazidime + CLA |
| Aztreonam |
| Aztreonam + CLA |